“Bioinformatics is still in its early stages, but its potential is enormous. It will play a key role in developing new drugs, diagnosing diseases and understanding the very nature of life.”
- Mark Zuckerberg
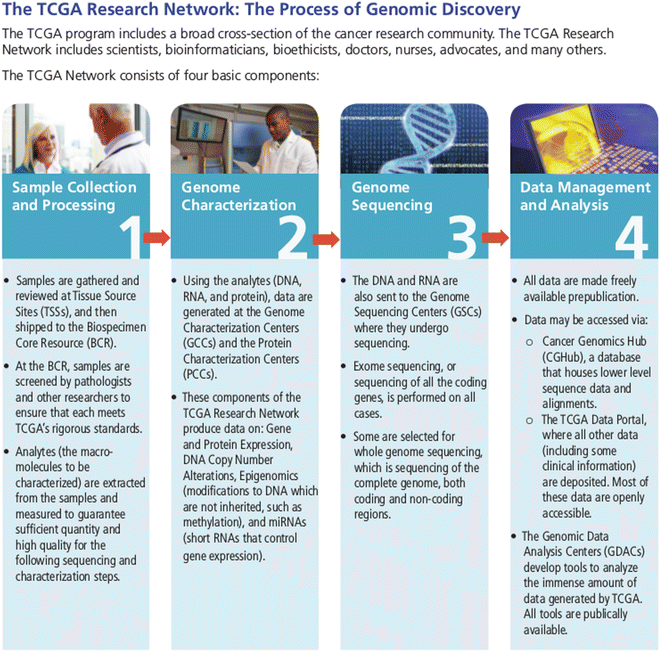
Introduction
TCGA, a groundbreaking collaborative effort initiated by the National Cancer Institute (NCI) and the National Human Genome Research Institute (NHGRI) in the United States, was designed to comprehensively characterize genomic alterations in diverse cancer types. The primary objectives were to advance our understanding of the molecular underpinnings of cancer, pinpoint potential therapeutic targets, and enhance cancer diagnosis and treatment. Commencing in 2005, the project concluded its data generation phase in 2013. TCGA encompassed the collection and analysis of genomic data from numerous cancer patients across a variety of cancer types, including, but not limited to, breast, lung, ovarian, colorectal, and brain cancers. The Cancer Genome Atlas (TCGA) involved a systematic and multi-step process to comprehensively characterize the genomic alterations in various types of cancer.
Here is an overview of the key steps in the TCGA process:
Sample collection
The primary objective of TCGA was to ensure a comprehensive representation of genetic diversity within each type of cancer. To achieve this, the collection of tumor tissue samples was undertaken, along with corresponding normal tissue samples whenever feasible. This meticulous process occurred predominantly during surgical procedures or biopsies, with explicit consent obtained from individuals diagnosed with cancer. The careful and deliberate sampling strategy was instrumental in capturing the intricate genetic nuances inherent in both cancerous and normal tissues.
DNA & RNA extraction
Genomic DNA, containing an organism’s entire set of genes, was meticulously isolated, serving as the foundation for comprehensive analyses. These analyses included whole-genome sequencing to identify genetic mutations and alterations, as well as DNA copy number analysis to assess variations in specific DNA segments, revealing insights into genomic instability often observed in cancer. Concurrently, RNA, which is responsible for transferring genetic information from DNA for protein synthesis, was also extracted. RNA played a crucial role in gene expression profiling, providing insights into active genes and variations in their activity levels between cancerous and normal tissues. This detailed molecular information proved pivotal for understanding the intricate molecular mechanisms underlying cancer development and progression.
Genomic analysis
1. Whole genome sequencing
Whole-genome sequencing was employed to scrutinize the genomic DNA comprehensively, aiming to discern genetic alterations encompassing mutations, copy number variations, and structural rearrangements. This crucial step sought to furnish a thorough and inclusive depiction of the genetic landscape inherent in the cancer samples.
2. RNA sequencing
The process of RNA sequencing was executed to quantify gene expression levels. This pivotal stage served the purpose of identifying fluctuations in gene expression, providing crucial insights into how genetic alterations influence the activity of specific genes and pathways. This analytical step played a vital role in unraveling the intricate dynamics of gene regulation associated with cancer development and progression.
Epigenetic analysis
TCGA’s exploration of epigenetic modifications involved a detailed examination of DNA methylation patterns, focusing on the addition of methyl groups to cytosine bases. This process aimed to uncover how changes in DNA methylation influence gene expression, playing a crucial role in cancer development. The analysis targeted the identification of specific methylation signatures associated with various cancer types, offering insights into the epigenetic landscape and molecular heterogeneity in cancer. These findings not only served as diagnostic markers but also provided potential targets for the development of precision therapies tailored to individual patients’ unique epigenetic profiles. TCGA’s epigenetic analysis significantly advanced our understanding of the intricate interplay between genetics and epigenetics in cancer biology.
Proteomic and clinical data collection
The integration of proteomic data within TCGA signified a holistic approach, providing a deeper understanding of the molecular landscape by analyzing protein expression levels. Proteins, being the effectors of biological functions, offer a dynamic perspective on cellular activity and contribute valuable information to complement the genomic findings. Additionally, the incorporation of comprehensive clinical information added contextual depth to the dataset, encompassing diverse aspects such as patient characteristics, treatment responses, and survival outcomes. This amalgamation of proteomic and clinical data facilitated a more nuanced exploration of the intricate relationship between molecular alterations and clinical phenotypes in cancer.
DATA – Integration, Analysis, Sharing and Accessibility
The vast amount of data generated from genomic, epigenomic, proteomic, and clinical analyses were integrated into a comprehensive dataset. Bioinformatics tools and computational methods were employed to analyze and interpret the complex relationships between genetic alterations and clinical characteristics. TCGA adopted an open-access philosophy, making its data publicly available to the research community. This approach facilitated collaboration and allowed researchers worldwide to explore, analyze, and build upon the TCGA dataset.
Bioinformatics support
Acknowledging the intricacies involved in the analysis of extensive genomic datasets, TCGA strategically instituted Genomic Data Analysis Centers (GDACs) as integral components of its framework. These specialized centers were assigned the crucial role of developing and implementing advanced bioinformatics tools and pipelines tailored specifically for processing and analyzing the substantial volume of genomic data generated throughout the project. The primary objectives of GDACs encompassed a diverse range of responsibilities.
Firstly, they assumed a pivotal role in upholding the quality, consistency, and reliability of the genomic data. This encompassed the implementation of rigorous quality control measures, addressing technical variations, and harmonizing diverse datasets to preserve the coherence of the information gathered from varied sources.
Secondly, GDACs were instrumental in spearheading the development of sophisticated bioinformatics tools and analytical pipelines. Crafted to navigate the complexities inherent in diverse genomic datasets, these tools enabled researchers to distill meaningful insights from the abundant information supplied by TCGA. The pipelines spanned various stages, from initial raw data preprocessing to advanced statistical analyses, thereby facilitating the identification of genetic alterations, biomarkers, and potential therapeutic targets.
In addition, GDACs played an active role in disseminating knowledge and expertise throughout the research community. Through engagement in training and collaboration initiatives, they ensured that researchers globally could adeptly navigate and leverage the TCGA data for their own investigations. This collaborative ethos significantly augmented the overall impact of TCGA, nurturing a community of researchers equipped with the indispensable tools and knowledge essential for advancing cancer genomics research.
Conclusion
In summary, the establishment of GDACs within TCGA underscored a commitment not merely to generating large-scale genomic data but also to providing the vital bioinformatics infrastructure necessary for transforming this data into meaningful and actionable insights within the realms of cancer research and personalized medicine. Findings from TCGA analyses were published in scientific journals, contributing to the broader understanding of cancer biology. The knowledge generated by TCGA has had a lasting impact on cancer research and clinical practice
– Shaistha Farheen. U. H
2nd year, BTech Biotechnology